The standard approach of patchwork is to place plots next to each other based
on the provided layout. However, it may sometimes be beneficial to place one
or several plots or graphic elements freely on top or below another plot. The
inset_element() function provides a way to create such insets and gives you
full control over placement.
Usage
inset_element(
p,
left,
bottom,
right,
top,
align_to = "panel",
on_top = TRUE,
clip = TRUE,
ignore_tag = FALSE
)Arguments
- p
A grob, ggplot, patchwork, formula, raster, nativeRaster, or gt object to add as an inset
- left, bottom, right, top
numerics or units giving the location of the outer bounds. If given as numerics they will be converted to
npcunits.- align_to
Specifies what
left,bottom, etc should be relative to. Either'panel'(default),'plot', or'full'.- on_top
Logical. Should the inset be placed on top of the other plot or below (but above the background)?
- clip
Logical. Should clipping be performed on the inset?
- ignore_tag
Logical. Should autotagging ignore the inset?
Examples
library(ggplot2)
p1 <- ggplot(mtcars) + geom_point(aes(mpg, disp))
p2 <- ggplot(mtcars) + geom_boxplot(aes(gear, disp, group = gear))
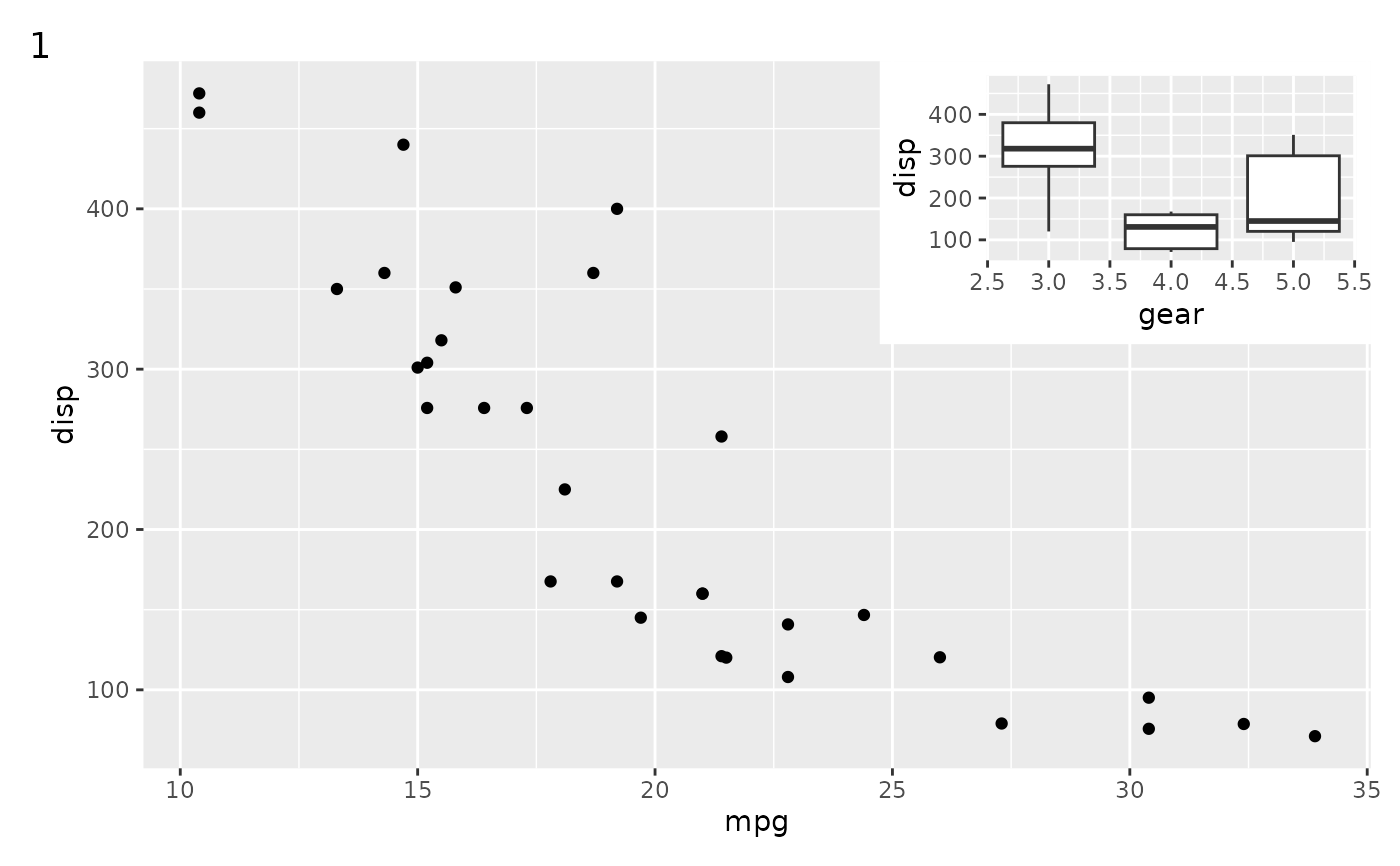
# Basic use
p1 + inset_element(p2, 0.6, 0.6, 1, 1)
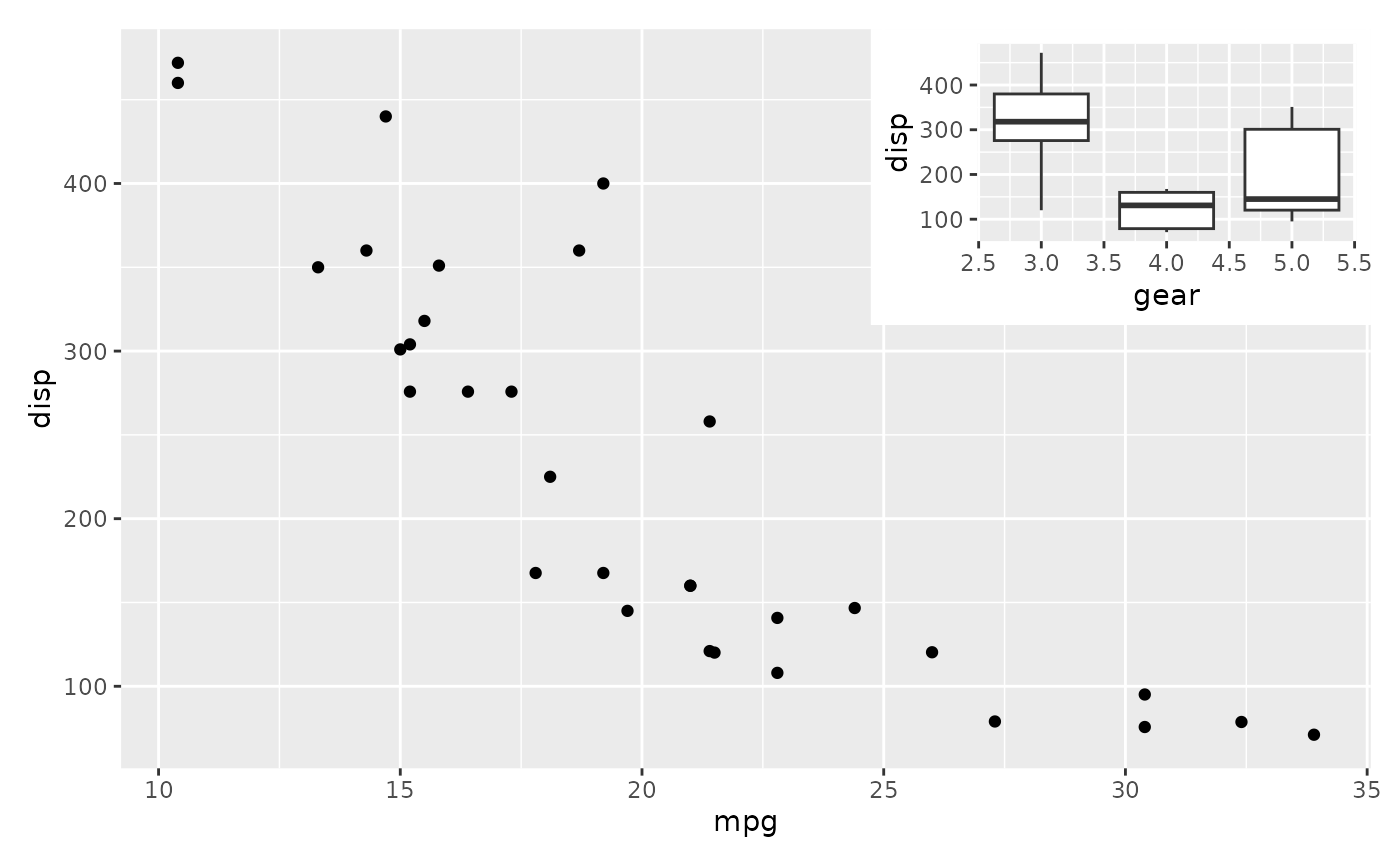 # Align to the full area instead
p1 + inset_element(p2, 0, 0.6, 0.4, 1, align_to = 'full')
# Align to the full area instead
p1 + inset_element(p2, 0, 0.6, 0.4, 1, align_to = 'full')
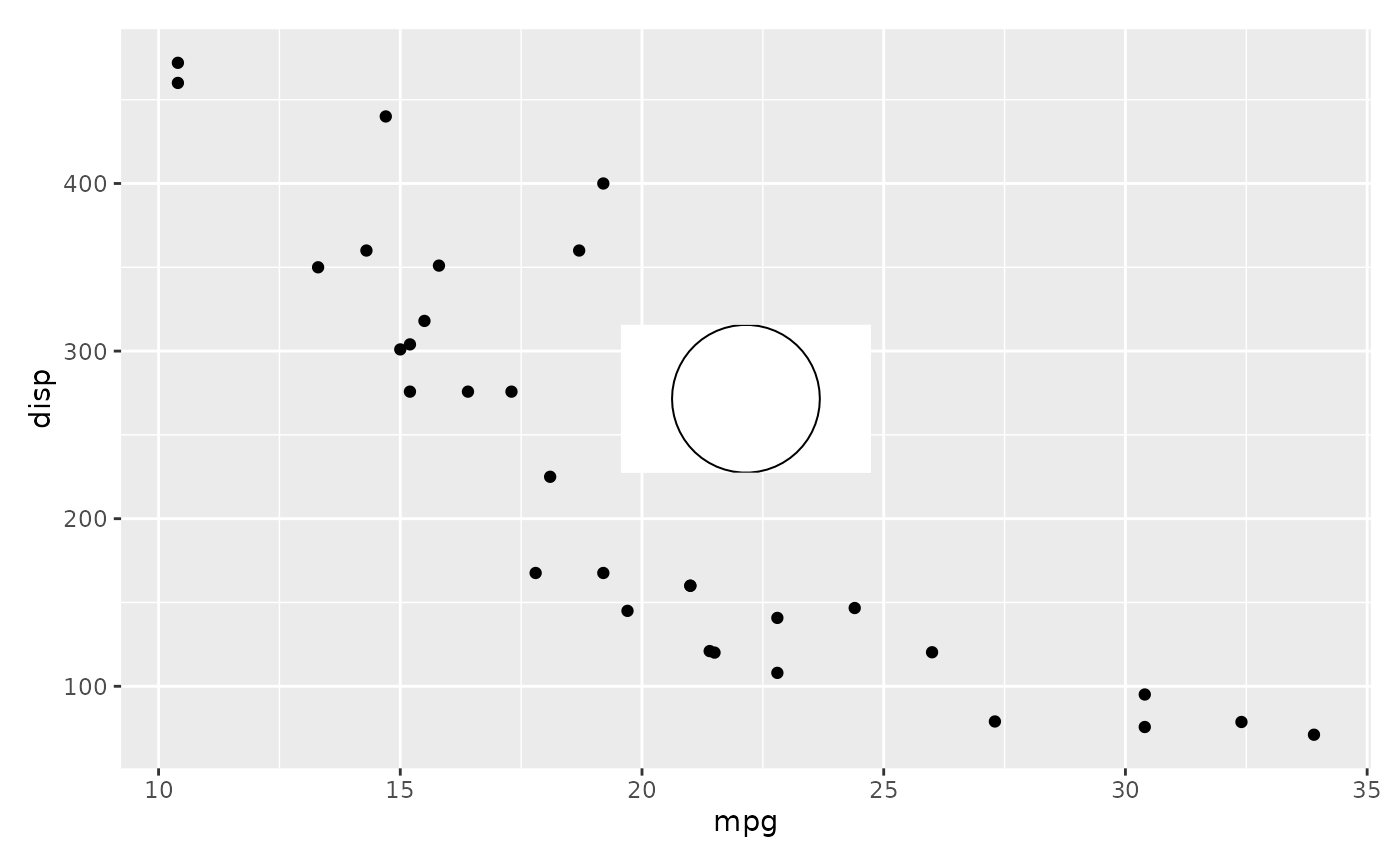
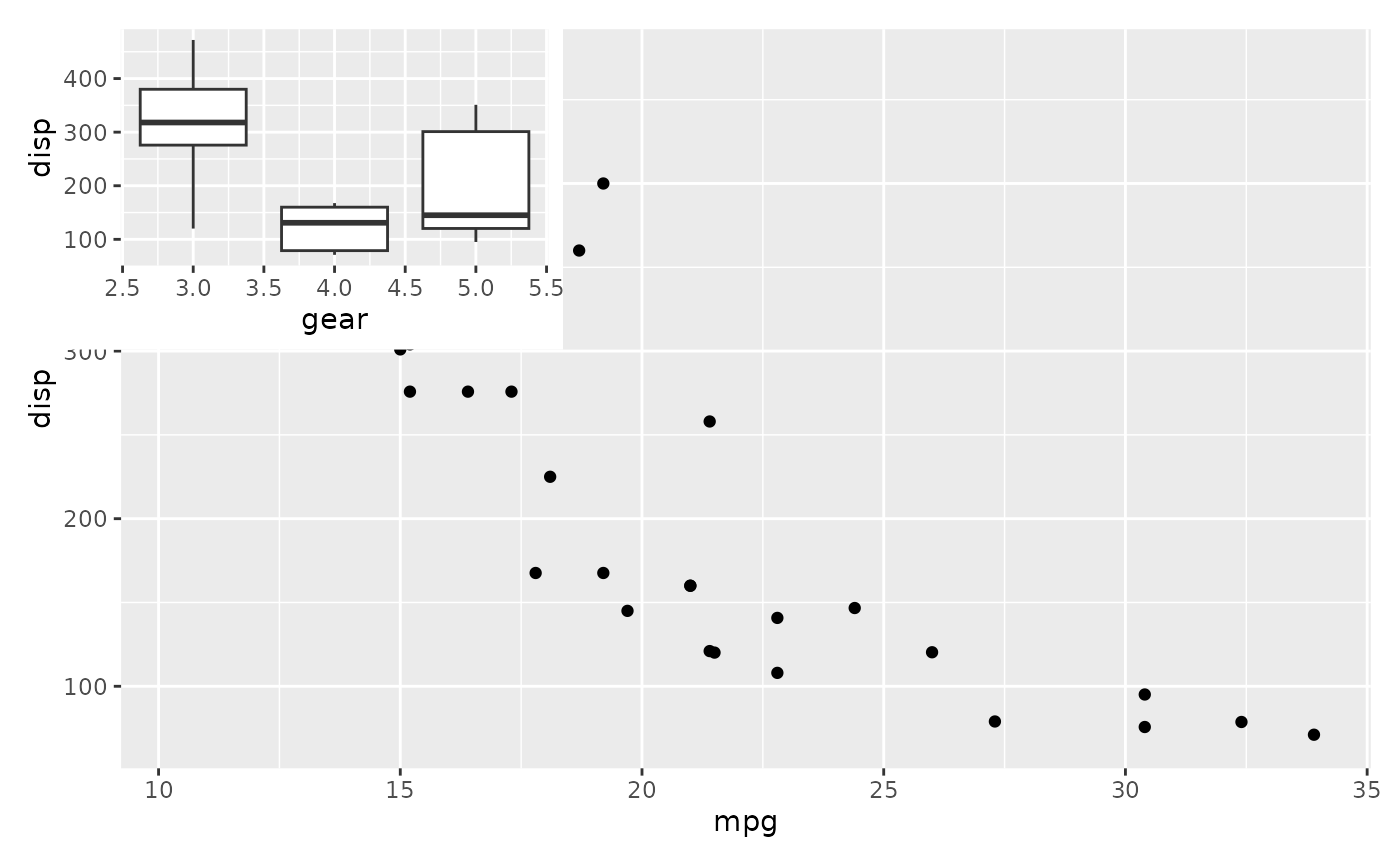 # Grobs and other objects can be added as insets as well
p1 + inset_element(grid::circleGrob(), 0.4, 0.4, 0.6, 0.6)
# Grobs and other objects can be added as insets as well
p1 + inset_element(grid::circleGrob(), 0.4, 0.4, 0.6, 0.6)
 if (requireNamespace('png', quietly = TRUE)) {
logo <- system.file('help', 'figures', 'logo.png', package = 'patchwork')
logo <- png::readPNG(logo, native = TRUE)
p1 + inset_element(logo, 0.8, 0.8, 1, 1, align_to = 'full')
}
if (requireNamespace('png', quietly = TRUE)) {
logo <- system.file('help', 'figures', 'logo.png', package = 'patchwork')
logo <- png::readPNG(logo, native = TRUE)
p1 + inset_element(logo, 0.8, 0.8, 1, 1, align_to = 'full')
}
 # Just as expected insets are still amenable to changes after the fact
p1 +
inset_element(p2, 0.6, 0.6, 1, 1) +
theme_classic()
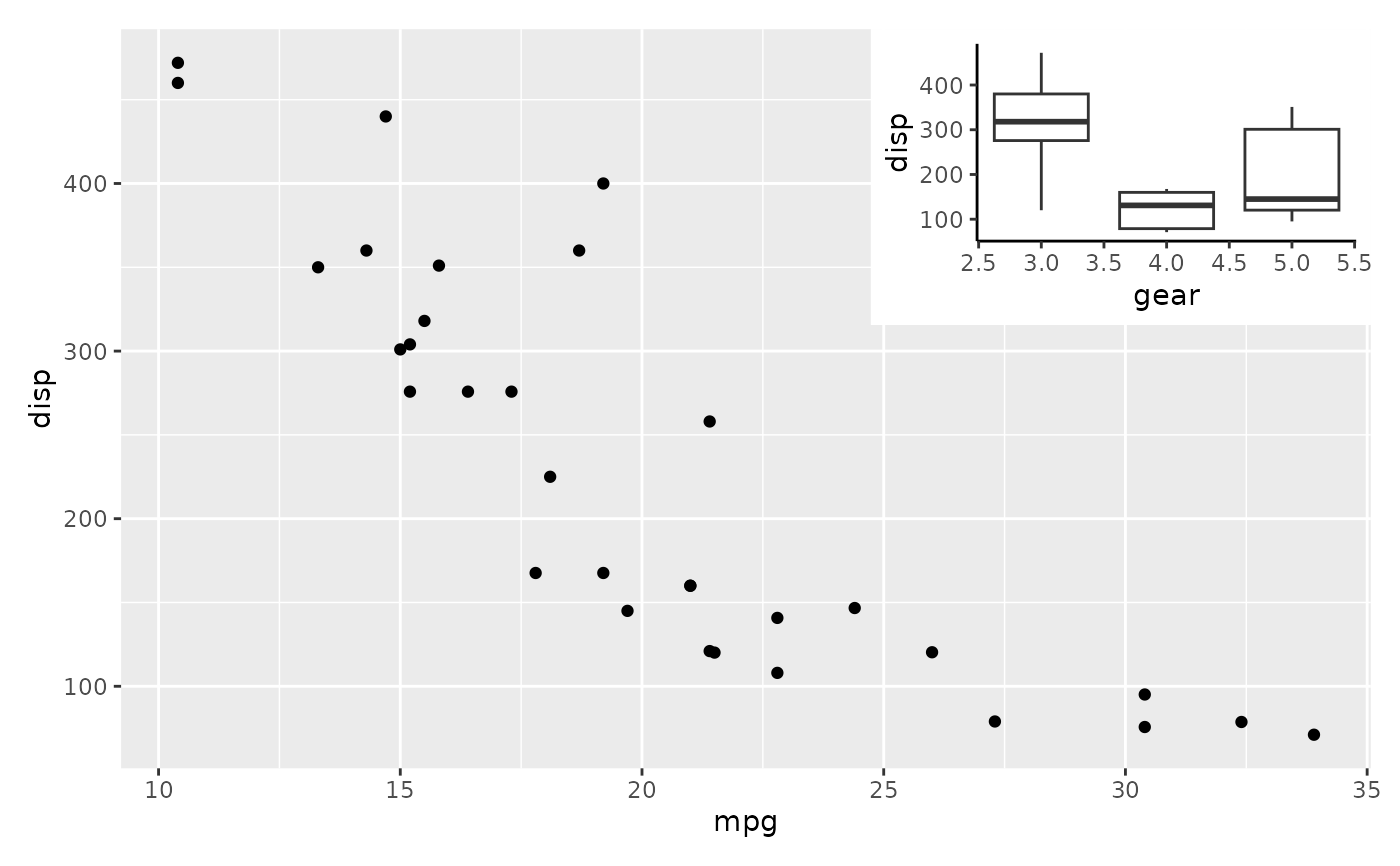
# Just as expected insets are still amenable to changes after the fact
p1 +
inset_element(p2, 0.6, 0.6, 1, 1) +
theme_classic()
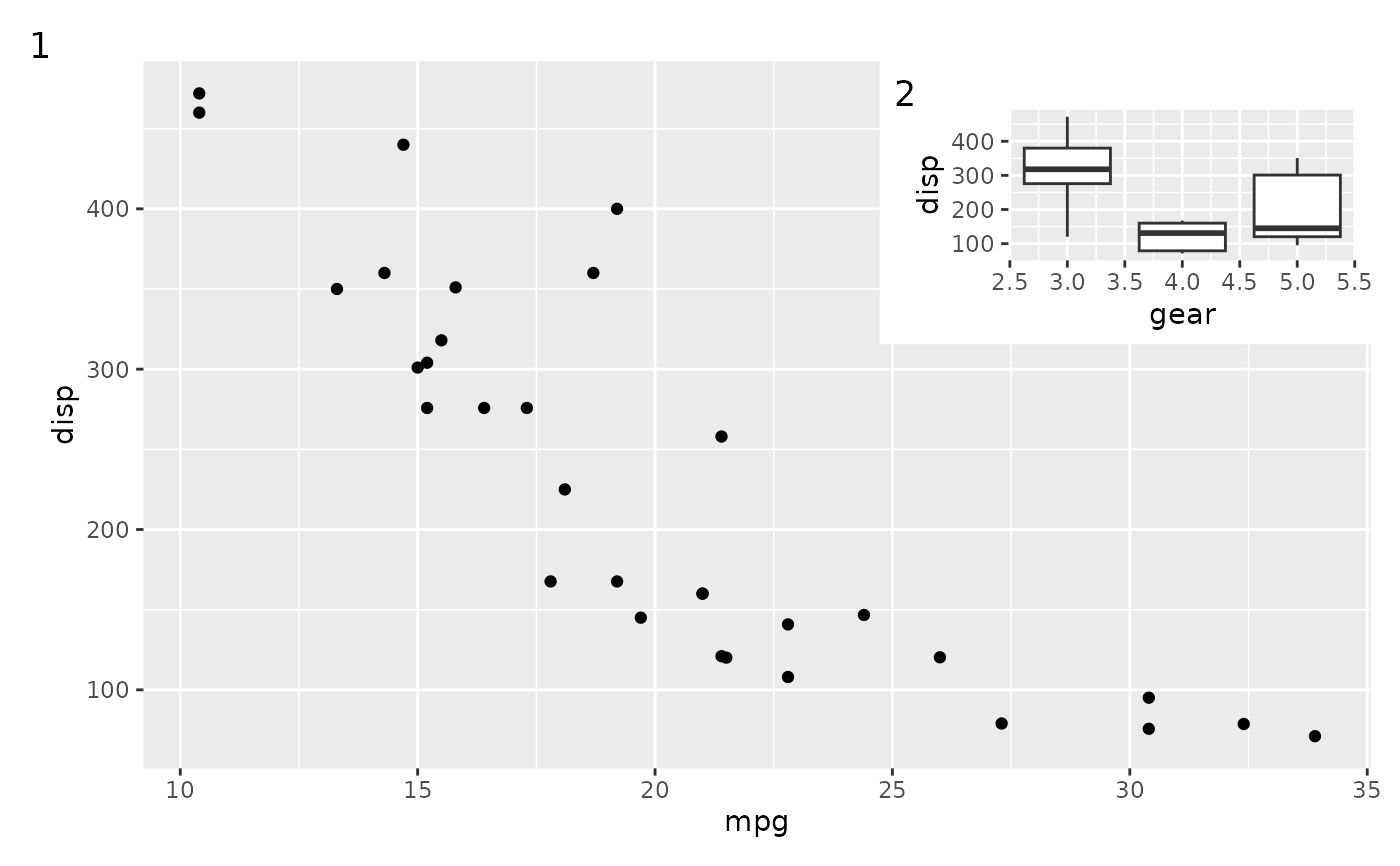
 # Tagging also continues to work as expected
p1 +
inset_element(p2, 0.6, 0.6, 1, 1) +
plot_annotation(tag_levels = '1')
# Tagging also continues to work as expected
p1 +
inset_element(p2, 0.6, 0.6, 1, 1) +
plot_annotation(tag_levels = '1')
 # but can be turned off, like for wrapped plots
p1 +
inset_element(p2, 0.6, 0.6, 1, 1, ignore_tag = TRUE) +
plot_annotation(tag_levels = '1')
# but can be turned off, like for wrapped plots
p1 +
inset_element(p2, 0.6, 0.6, 1, 1, ignore_tag = TRUE) +
plot_annotation(tag_levels = '1')